–an epigenetic clock. Implementing a checkpoint on the road to healthier living
Work Flow:
•Data from EWAS repository (Other databases – European Genome-Phenome Archive (UK population)…)
•Database created in MySQL Workbench
•metadata (sample information), beta_values (CpG sites and the probabilities of that particular site being methylated in the DNA in that sample – Beta values), molecular_clocks (biological clocks, their weights, etc), epigenetic_age (predicted epigenetic age)
•metadata_additional – there are many other variables that could be incorporated in the database as well (age and bmi)
•EPIC array denoted as 850K; N/A’s replaced with 0’s for ease of calculation later; about 16000+ CpG sites per sample
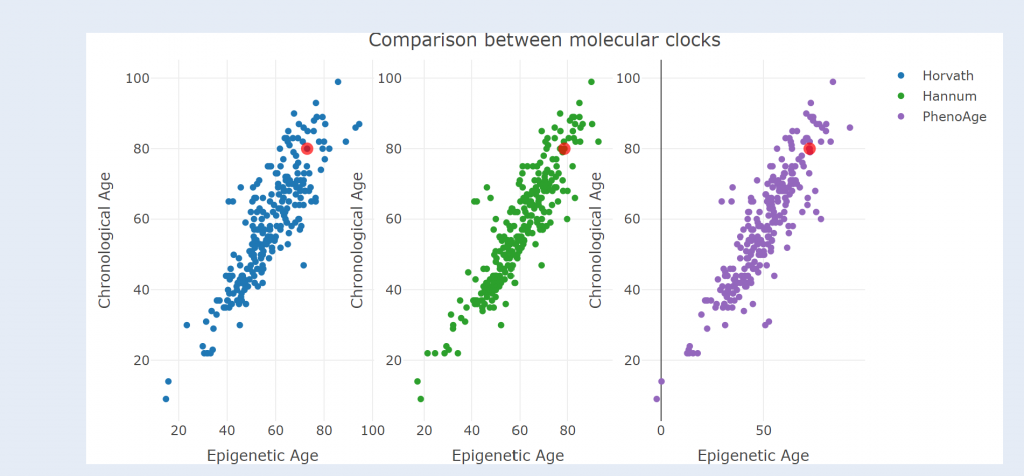
•SMART app (for DNA methylation analysis and visualization)
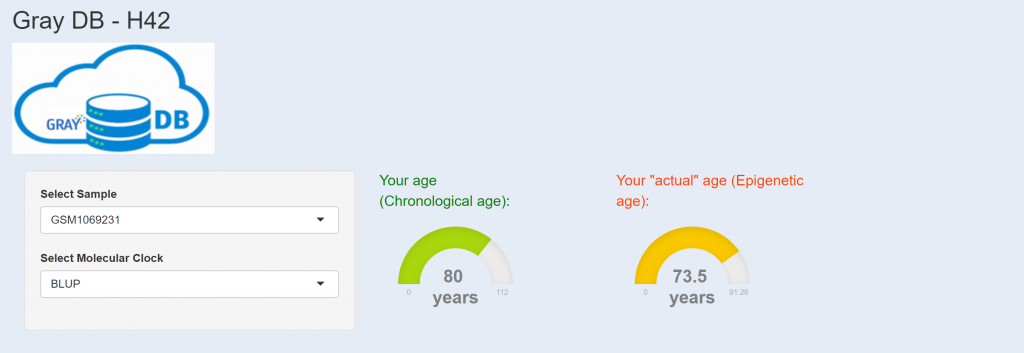
–Look at your epigenetic age at here: